Understanding how cancer changes the way genes are expressed is critical to developing better diagnostics and therapies. RNA sequencing (RNA-Seq) has become a cornerstone of cancer research because it offers an unbiased, high-resolution view of the transcriptome, revealing gene expression shifts, alternative splicing patterns, fusion transcripts, and non-coding RNA dynamics that other methods often miss.
Unlike traditional microarrays, RNA-Seq can uncover previously unknown transcripts and molecular signatures, making it especially valuable in studying tumours with high genetic variability. Yet despite its power, many researchers face challenges in interpreting complex datasets and integrating RNA-Seq results with other omics layers.
In this post, we’ll explore the key applications of RNA-Seq in cancer research, from tumour subtyping and biomarker discovery to therapy response prediction. We’ll also highlight both the technology’s impact and limitations as it moves closer to clinical translation.
TL;DR
- RNA-Seq enables comprehensive, unbiased transcriptome profiling in cancer, supporting detection of gene expression changes, alternative splicing, and novel transcripts across tumor types.
- Advances from next-generation to third-generation sequencing have improved isoform resolution and identification of gene fusions, addressing complex transcriptomic features in cancer.
- Single-cell and spatial transcriptomics provide high-resolution insights into tumor heterogeneity, microenvironmental interactions, and mechanisms of therapy resistance.
- RNA-Seq is integral to biomarker discovery, therapy selection, drug development, and real-time monitoring through liquid biopsy applications, but data integration and standardization remain significant challenges.
- Ongoing innovation in sequencing technologies and analytical methods is essential to overcome technical and clinical barriers, enabling more effective translation of RNA-Seq findings into oncology practice.
What is RNA Sequencing
RNA sequencing (RNA-Seq) is a powerful laboratory technique used to analyze the quantity and sequences of RNA molecules present in a biological sample, providing a comprehensive view of gene expression and the transcriptome, the complete set of RNA transcripts produced by the genome under specific circumstances or in a specific cell.
What Does RNA-Seq Reveal?
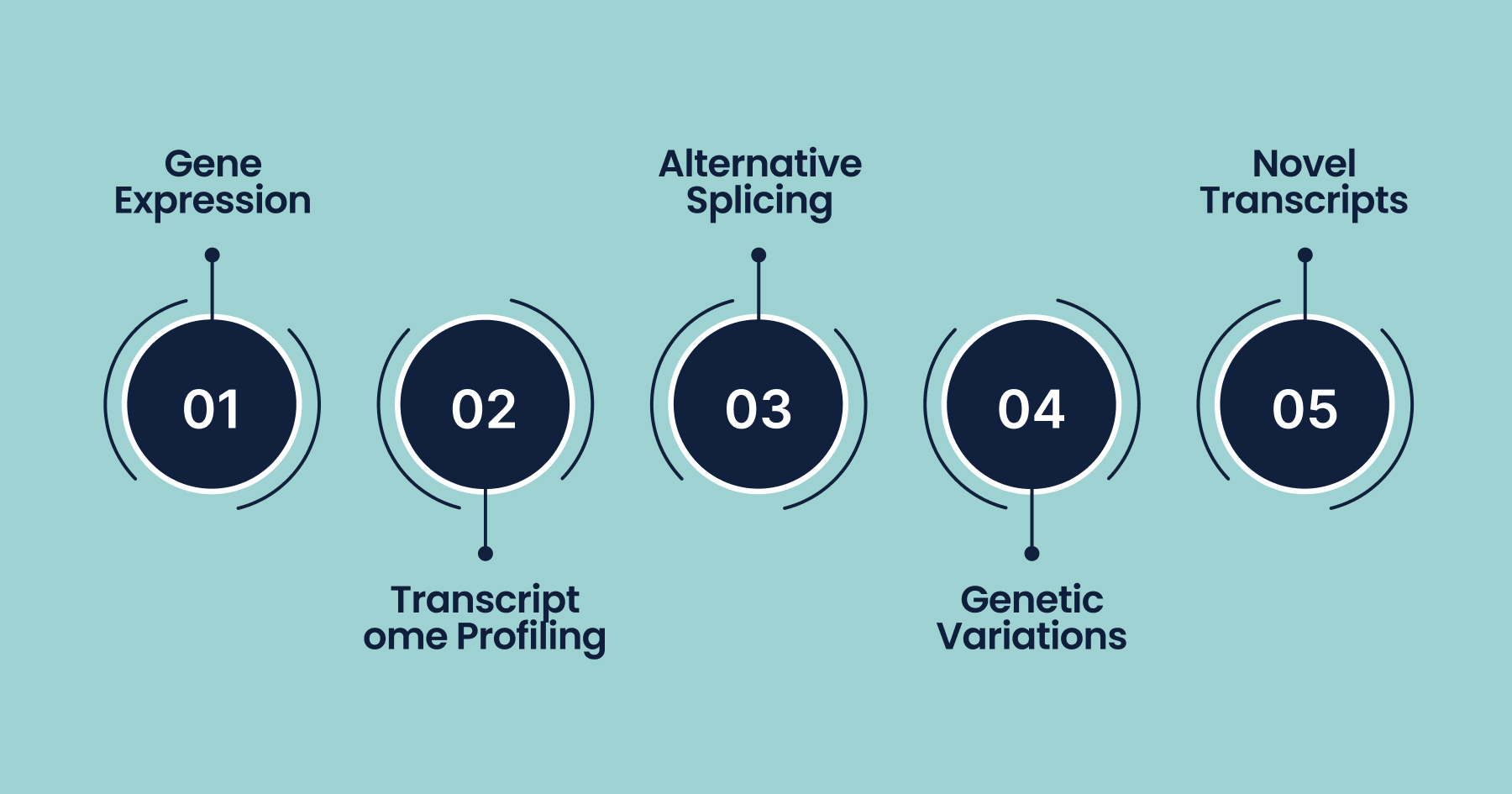
- Gene Expression: Identifies which genes are active (expressed) in the sample and quantifies their expression levels.
- Transcriptome Profiling: Provides a snapshot of all RNA molecules present, including messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and various non-coding RNAs.
- Alternative Splicing & Isoforms: Detects different splice variants and transcript isoforms from the same gene.
- Genetic Variations: Identifies mutations, single-nucleotide polymorphisms (SNPs), gene fusions, and other sequence variations.
- Novel Transcripts: Discovers previously unannotated RNA molecules and gene boundaries
Now, let’s find out about the development of next-generation sequencing in cancer research.
Development From First Generation to Next Generation Sequencing in Cancer Research
In cancer research, RNA-Seq technology provides unprecedented insights into how malignant cells differ from healthy tissue at the molecular level.
Next-generation sequencing (NGS)
First-generation sequencing (Sanger method) provided foundational insights into RNA structure but faced limitations in throughput and cost. The evolution from first-generation Sanger sequencing to next-generation sequencing (NGS) marked a paradigm shift in cancer genomics.
- Platforms like Illumina revolutionized RNA-Seq by enabling high-throughput, cost-effective transcriptome profiling.
- Facilitated large-scale cancer studies such as The Cancer Genome Atlas (TCGA)
- Allowed identification of subtype-specific gene expression signatures and driver mutations
Emergence of Third-Generation Sequencing and Its Impact
Third-generation technologies address NGS limitations in resolving complex isoforms.
- Include PacBio SMRT and Oxford Nanopore long-read sequencing
- Capture full-length transcripts, crucial for detecting alternative splicing and gene fusions
- Improve detection of complex isoforms common in cancers (e.g., TMPRSS2-ERG in prostate cancer, BCR-ABL1 in leukemia)
- Identify more cancer-specific isoforms compared to short-read methods.
- Higher raw error rates than NGS, but bioinformatics corrections now achieve >99% accuracy
Role of Single-Cell and Spatial Transcriptomics in Cancer
scRNA-seq allows transcriptomic profiling at the resolution of individual cells, revealing the heterogeneity within tumors that bulk sequencing cannot capture.
- Single-cell RNA-Seq profiles gene expression at the individual cell level
- Reveals tumor heterogeneity and rare resistant subpopulations (e.g., therapy-resistant melanoma cells)
- Spatial transcriptomics maps RNA expression within the spatial context of tumor tissue (e.g., 10x Visium, MERFISH)
- Provides insights into tumor microenvironment and immune cell localization
- Recent studies have highlighted immune exclusion zones contributing to therapy resistance in pancreatic cancer
- Applications include studying metastasis, stromal interactions, and clonal evolution in tumors.
For cancer researchers, selecting the right tool depends on the biological question, whether prioritizing isoform resolution (long-read), cellular diversity (single-cell), or tissue architecture (spatial). Frameworks like Seurat and Scanpy are widely adopted for integration, but standardization is ongoing.
As you design experiments, consider hybrid approaches: use NGS for broad discovery, long-read for structural validation, and spatial/scRNA-seq for mechanistic depth. Each of these technologies contributes a unique layer of biological resolution—when integrated thoughtfully, they bring us closer to decoding the full complexity of cancer transcriptomes.
With these powerful tools in hand, scientists are now applying RNA-Seq across a range of cancer types, uncovering disease-specific molecular signatures and therapeutic opportunities.
RNA-Seq Applications in Cancer Research
RNA-Seq has revealed distinct molecular subtypes, resistance mechanisms, and evolutionary trajectories across various cancer types.
- Breast Cancer Research
Breast cancer research has benefited enormously from RNA-seq applications.
- Scientists have identified distinct molecular subtypes: luminal A, luminal B, and HER2-enriched.
- A groundbreaking 2023 study published in Nature Cancer used single-cell RNA-seq to map the cellular ecosystem of triple-negative breast cancer (TNBC), revealing previously unknown interactions between cancer cells and immune infiltrates.
- Researchers identified specific T-cell exhaustion signatures that predict immunotherapy response, leading to improved patient selection for checkpoint inhibitor treatments.
- The METABRIC consortium’s recent work has demonstrated how RNA-seq can predict long-term survival outcomes with greater accuracy than traditional clinical parameters.
These molecular signatures are now being integrated into clinical decision-making tools, helping oncologists tailor treatment strategies based on individual tumor biology.
- Lung Cancer Insights
RNA-seq applications in lung cancer research have yielded remarkable insights into treatment resistance mechanisms.
- The recent TRACERx study, published in Nature in 2023, used multi-region RNA sequencing to track how lung tumors evolve during treatment. This reveals that therapeutic resistance often emerges from pre-existing rare cell populations rather than new mutations.
- Scientists have discovered that non-small cell lung cancer (NSCLC) patients with high expression of interferon-gamma signaling genes show significantly better responses to PD-1/PD-L1 inhibitors. This finding has led to the development of predictive biomarker panels for lung cancer, now used in clinical practice.
- RNA-seq analysis revealed that tumors developing resistance to third-generation EGFR inhibitors activate alternative signaling pathways, including MET amplification and epithelial-to-mesenchymal transition programs. This discovery has informed combination therapy strategies that prevent resistance for lung cancer before it emerges.
- Colorectal Cancer Analysis
The Colorectal Cancer Subtyping Consortium’s recent work has used RNA-seq to refine the consensus molecular subtypes (CMS) classification system.
- Their 2024 study in Cell identified additional molecular features that predict response to immunotherapy, particularly in microsatellite stable tumors traditionally considered immune-cold.
- Revolutionary research has shown how RNA-seq can track the molecular evolution from normal colon epithelium through adenoma to invasive carcinoma.
- Scientists identified specific transcriptional checkpoints where interventions might prevent cancer progression, opening new avenues for chemoprevention strategies.
- Leukemia and Hematologic Malignancies
RNA-seq has significantly advanced clinical oncology and the understanding of acute myeloid leukemia (AML) heterogeneity.
- The Beat AML consortium’s recent findings demonstrated that RNA-seq-based classification predicts treatment response more accurately than traditional cytogenetics alone.
- Researchers have identified minimal residual disease signatures using RNA-seq that can detect one leukemic cell among 100,000 normal cells, enabling early relapse prediction.
- This ultrasensitive detection capability is advancing treatment monitoring and relapse prediction in acute lymphoblastic leukemia patients.
These insights across cancer types are now driving the development of advanced biomarker panels and personalised therapeutic strategies.
Biomarker Identification Techniques
The power of RNA-Seq lies in its ability to uncover robust molecular signatures that serve as biomarkers for diagnosis, prognosis, and therapeutic response.
- Multi-Modal Integration Approaches
Recent advances in biomarker discovery combine RNA-seq with other omics technologies to create comprehensive molecular portraits. The Pan-Cancer Analysis of Whole Genomes project has integrated RNA-seq with whole-genome sequencing data from over 2,600 cancer samples, revealing how structural variants influence gene expression and identifying new therapeutic targets.
Scientists are now using proteogenomics approaches that combine RNA-seq with mass spectrometry-based proteomics to identify tumor-specific neoantigens for personalized cancer vaccines. This integrated approach has shown remarkable success in melanoma and renal cell carcinoma patients.
- Liquid Biopsy Applications
RNA-seq analysis of circulating tumor cells (CTCs) and extracellular vesicles represents a rapidly advancing field. Recent studies have demonstrated the ability to detect cancer recurrence months before conventional imaging through RNA-seq analysis of blood samples.
The GRAIL Galleri test, which uses next-generation sequencing of cell-free DNA, primarily through methylation profiling, to detect over 50 cancer types from a single blood draw. Clinical trials are currently evaluating its potential for population-wide cancer screening.
- Spatial Transcriptomics in Understanding Cancer Architecture
A 2024 study in Cell used spatial RNA-seq to reveal how immune cells organize around tumor margins, identifying spatial gene expression patterns that predict immunotherapy response. This work has practical implications for pathologists who can now incorporate spatial molecular information into diagnostic workflows.
This spatial perspective is particularly valuable for predicting and improving responses to immunotherapies, as it reveals how immune cells and tumor cells interact within their native environments.
Immunotherapy and Treatment Response Prediction
By distinguishing between functional and exhausted immune cell populations, RNA-Seq guides patient selection, optimizes therapeutic strategies, and enhances the effectiveness of emerging immunotherapies.
- Checkpoint Inhibitor Response Signatures
RNA-seq has identified robust gene expression signatures that predict response to immune checkpoint inhibitors across multiple cancer types. The tumor inflammation signature (TIS), developed through extensive RNA-seq studies, measures the level of adaptive immune infiltration and has shown consistent predictive value across clinical trials.
Recent work has revealed that the effectiveness of checkpoint inhibitors depends not just on immune infiltration but also on the functional state of tumor-infiltrating lymphocytes.
RNA-seq analysis can distinguish between functional and exhausted T-cell populations, providing more nuanced predictions of treatment response.
- CAR-T Cell Therapy Optimization
RNA-seq advances CAR-T cell therapy development and monitoring. Scientists use single-cell RNA-seq to track how CAR-T cells evolve after infusion, identifying molecular markers associated with persistence and anti-tumor efficacy.
Recent studies have shown that Pre-infusion RNA-Seq profiling of CAR-T cells helps predict their persistence and therapeutic potential, allowing for quality control measures that improve patient responses.
Beyond diagnostics, RNA-Seq plays a growing role in drug discovery—helping identify novel targets, understand resistance mechanisms, and repurpose existing therapies based on transcriptomic signatures.
Drug Discovery and Development Applications
The integration of RNA-Seq into drug discovery pipelines has accelerated the identification and validation of new drug targets, uncovered synthetic lethal interactions, and enabled drug repurposing efforts. These are:
- Target Identification and Validation
RNA-seq has accelerated cancer drug discovery by identifying previously unknown therapeutic targets. The Cancer Dependency Map project uses RNA-seq to systematically identify genes essential for cancer cell survival across hundreds of cell lines, revealing potential drug targets with unprecedented scope.
Recent breakthroughs include the identification of synthetic lethal interactions through RNA-seq analysis. Scientists have discovered that cancers with specific gene expression profiles become dependent on particular metabolic pathways, creating opportunities for targeted metabolic therapies.
- Drug Repurposing Opportunities
RNA-seq databases are being mined to identify existing drugs that might be repurposed for cancer treatment. The Connectivity Map project uses RNA-seq signatures to match disease states with drug effects, leading to clinical trials of unexpected drug combinations.
A notable success story involves the discovery that cardiac glycosides, traditionally used for heart conditions, show anti-cancer activity against specific RNA-seq-defined tumor subtypes. This finding has led to clinical trials exploring digitoxin in cancer treatment.
These advances in drug development are now being translated into clinical practice, where RNA-Seq is at the forefront of precision medicine and innovative treatment strategies.
RNA-Seq in Cancer Treatment: Latest Breakthroughs
Recent breakthroughs in RNA-Seq have transformed clinical oncology, enabling precision medicine approaches that match patients with targeted therapies based on their tumor’s molecular profile. Here are some of the latest breakthroughs on RNA-seq applications in cancer research
- Precision Medicine Applications
RNA-seq enables precision medicine by identifying the specific molecular alterations driving each patient’s cancer. Recent clinical trials have demonstrated the effectiveness of this approach. The NCI-MATCH trial uses comprehensive molecular profiling, including RNA-seq, to match patients with targeted therapies based on their tumor’s genetic characteristics rather than cancer type.
- Immunotherapy Selection
RNA-seq has advanced immunotherapy selection by identifying tumors likely to respond to checkpoint inhibitors. Scientists measure immune gene expression signatures, tumor mutational burden, and microsatellite instability status to predict treatment response.
Recent breakthrough research has used RNA-seq to identify “hot” tumors with high immune infiltration versus “cold” tumors that may require combination therapies to stimulate immune responses.
- Drug Resistance Mechanisms
Researchers use RNA-seq to understand how cancers develop resistance to targeted therapies. By comparing pre-treatment and post-progression samples, scientists identify the molecular changes that allow cancer cells to survive treatment.
This research has led to combination therapy strategies that prevent or overcome resistance. For example, studies of EGFR-mutant lung cancers have revealed multiple resistance pathways, leading to sequential treatment strategies that target different molecular mechanisms.
- Monitoring Treatment Response
RNA-seq analysis of circulating tumor cells (CTCs) and extracellular RNA, along with DNA-based analysis of cell-free DNA (cfDNA), enables real-time monitoring of treatment response without invasive biopsies. This liquid biopsy approach enables oncologists to adjust treatment strategies based on molecular changes occurring in patients’ tumors.
Despite these remarkable achievements, several technical, computational, and clinical challenges remain, highlighting the need for continued innovation and collaboration in the field.
Current Limitations and Future Directions
RNA-seq faces technical, computational, and clinical challenges, including issues with sample quality, data standardization, and integration with other data types. To fully realize the promise of RNA-seq in cancer research and patient care, continued innovation in sequencing technologies, bioinformatics, and regulatory frameworks will be required.
- Technical Challenges
Despite remarkable advances, RNA-seq applications in cancer research face several limitations. Sample quality remains a significant challenge, particularly for formalin-fixed, paraffin-embedded (FFPE) tissues commonly used in clinical settings. RNA degradation can compromise results and limit the sensitivity of biomarker detection.
Standardization across laboratories presents another hurdle. Different sample preparation methods, sequencing platforms, and analysis pipelines can produce varying results, making it difficult to compare studies or implement findings clinically.
- Computational Bottlenecks
The massive datasets generated by RNA-seq require sophisticated computational infrastructure and expertise. Many research institutions lack the bioinformatics resources needed to fully exploit RNA-seq data, creating barriers to widespread adoption.
Data integration presents additional challenges. Combining RNA-seq results with other omics data, clinical information, and imaging studies requires advanced analytical approaches that are still being developed.
- Clinical Implementation Barriers
Translating RNA-seq research findings into clinical practice remains complex. Regulatory approval processes for genomic tests are lengthy and expensive, slowing the implementation of promising biomarkers.
Cost considerations also limit clinical adoption. While RNA-seq costs have decreased dramatically, comprehensive transcriptomic analysis remains expensive compared to traditional diagnostic tests.
As the field addresses these hurdles, the ongoing evolution of RNA-Seq technologies promises to further improve cancer research and patient care in the years ahead.
Biostate AI: The Complete Solution for RNA Sequencing
Biostate AI addresses these challenges by offering an end-to-end RNA sequencing platform that streamlines every step, from sample collection to final data insights. By automating laboratory processes and providing AI-driven analytics, we help researchers to focus on scientific discovery rather than technical and operational hurdles.
Key Features of Biostate AI
- Unbeatable Pricing: High-quality sequencing results starting at $80 per sample.
- Rapid Turnaround: Receive results in just 1–3 weeks.
- Complete Transcriptome Insights: Comprehensive RNA-Seq covering both mRNA and non-coding RNA.
- Minimal Sample Requirement: Process samples as small as 10µL blood, 10ng RNA, or 1 FFPE slide.
- Low RIN Compatibility: Compatible with RNA samples having RIN as low as 2 (versus the typical ≥5).
- OmicsWeb: The AI-Ready OmicsWeb is a comprehensive, intuitive platform combining robust data storage with automated analysis workflows, making your data ready for AI-driven insights. Features include:
- Multi-omics support (RNA-Seq, WGS, methylation, single-cell)
- AI Copilot for natural language data analysis
- Automated pipelines from raw data to publication-ready insights
- Disease Prognosis AI (Biobase): Transform RNA data into accurate disease predictions and therapy guidance using Biobase, with internal experiments showing:
- 89% accuracy in predicting drug toxicity in rodents
- 70% accuracy in therapy selection for AML
- Flexible Sample Compatibility: Works with blood, tissue, culture, and purified RNA samples for both long-term and point-in-time experiments.
Biostate AI empowers researchers with a unified, affordable, and robust RNA sequencing solution, delivering high-quality data and actionable insights to advance scientific discovery in cancer and beyond.
Final Words
RNA-seq applications in cancer research continue to yield transformative discoveries that are reshaping oncology. From identifying new therapeutic targets to predicting treatment responses, this technology provides the molecular insights necessary for advancing precision cancer medicine.
Biostate AI offers a truly cost-effective RNA sequencing solution, combining affordability with scientific rigor and flexibility. With pricing starting at $80 per sample, rapid turnaround, and compatibility with even low-integrity samples (RIN as low as 2), our platform makes advanced RNA-seq accessible to labs of all sizes.
Choose us to get high-quality data to support your broad range of research needs with our cost-effective solution. Request a personalized demo today!
FAQs
- How does RNA-seq differ from DNA sequencing in cancer research?
While DNA sequencing reveals the genetic blueprint, the permanent instructions encoded in our genes, RNA-seq captures the dynamic expression of those genes.
In cancer research, this distinction is crucial. A patient might carry a cancer-predisposing gene mutation (detected by DNA sequencing), but RNA-seq reveals whether that gene is actually active and influencing cancer development. This real-time molecular activity provides more immediate, actionable information for treatment decisions.
- What types of cancer samples can be analyzed using RNA-seq?
RNA-seq can analyze fresh or frozen tissue, FFPE blocks, blood, bone marrow, pleural or ascitic fluid, and single cells. This flexibility allows molecular profiling from routine clinical and archived samples.
This flexibility is particularly valuable for patients, as it means molecular analysis can be performed on samples collected through routine clinical procedures, minimizing the need for additional invasive procedures.
- How does RNA-seq differ from traditional genetic testing in cancer care?
While traditional genetic testing examines the DNA blueprint revealing what could potentially happen RNA-seq captures the dynamic reality of what is actively occurring within cancer cells. This distinction carries profound implications for patient care and treatment decisions. Traditional genetic testing might identify inherited mutations that increase cancer risk, but RNA-seq reveals whether those genetic changes are actively driving tumor growth.
