Choosing the right RNA sequencing method can impact your research. PacBio and Nanopore are two popular options, each with its own strengths. Both offer long-read sequencing, but they differ in accuracy, real-time sequencing, and cost.
If you’re working with RNA sequencing, knowing these differences can help you pick the best fit for your research. This blog will highlight the key points of both methods so you can make a choice based on what works best for your project.
PacBio: Long-Read Sequencing
PacBio’s Single Molecule Real-Time (SMRT) technology has transformed DNA sequencing with long, accurate reads that capture full-length DNA molecules. This capability allows researchers to explore complex genomic regions, identify repetitive sequences, and detect structural variations.
PacBio is ideal for de novo genome assembly, haplotype phasing, and epigenetic analysis, providing detailed insights into genes, regulatory elements, and genetic variations.
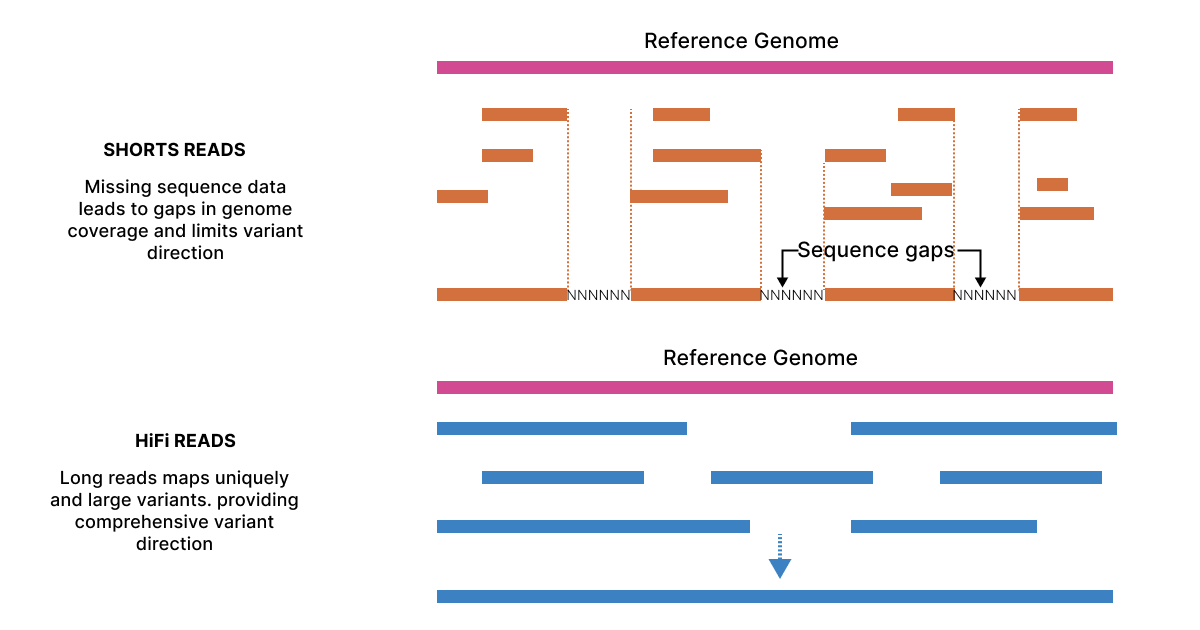
Recently, the introduction of HiFi sequencing builds on SMRT, offering highly accurate, full-length reads through circular consensus sequencing (CCS). This method improves sequencing precision, especially for complex regions and structural variations.
HiFi sequencing is essential for applications like rare variant detection, structural variant identification, and transcriptome analysis, providing reliable and complete genetic data. Its high accuracy and ability to handle complex genomic features make it a critical tool for advanced genetic research.
Nanopore: Real-Time Sequencing
Nanopore sequencing technology by Oxford Nanopore takes a different approach to DNA and RNA sequencing. Instead of amplifying nucleic acids, it passes them through a nanopore and measures changes in electrical current, offering real-time sequencing. One key advantage is its ability to sequence RNA directly, without the need for cDNA conversion, capturing modifications like methylation.
Nanopore sequencing is also portable, cost-effective, and scalable, making it suitable for both field research and large-scale studies. It’s commonly used for genomic assembly, transcriptome analysis, and epigenetic research, providing a deeper understanding of genetic variation and gene regulation. Plus, with long-read sequencing and immediate data feedback, it’s a powerful tool for a variety of genetic studies.
Comparison Between PacBio and Nanopore RNA Sequencing Technologies
RNA sequencing is key to understanding gene expression, splicing, and regulatory mechanisms. PacBio and Nanopore are two powerful RNA sequencing technologies that bring unique strengths to the table.
1. Sample Preparation
- PacBio: RNA is first converted into complementary DNA (cDNA) before sequencing. Though this step adds extra time, it ensures better stability and reproducibility. PacBio’s HiFi sequencing improves the quality of cDNA sequencing, making it especially useful for accurate and reliable RNA studies.
- Nanopore: Nanopore shines with its direct RNA sequencing, which bypasses the cDNA conversion. This approach preserves the RNA’s natural state, allowing for the detection of modifications like methylation. It’s ideal for studying gene regulation and other RNA modifications that traditional methods might miss.
2. Read Lengths and Accuracy
- PacBio: PacBio is known for producing long reads that span thousands of nucleotides, capturing entire transcripts. These long reads make it easier to study alternative splicing, isoforms, and complex RNA structures. With HiFi sequencing, PacBio ensures high accuracy, even with long sequences, offering confident results in RNA analysis, especially for rare variant detection.
- Nanopore: Nanopore also delivers long reads, allowing it to sequence RNA directly. However, the raw accuracy of Nanopore tends to be lower than PacBio, particularly for longer reads. The company has made progress in enhancing accuracy with base-calling algorithms, but PacBio still maintains a higher level of precision for critical RNA applications.
3. Direct Sequencing
- PacBio: PacBio does not offer direct RNA sequencing, as RNA must first be converted into cDNA. This step may add complexity but ensures greater stability, especially when working with challenging RNA samples. HiFi sequencing gives full-length, high-quality cDNA reads, making PacBio ideal for projects that need precision.
- Nanopore: One of Nanopore’s major strengths is its ability to perform direct RNA sequencing, which eliminates the need for cDNA conversion. This allows for a more accurate representation of the RNA molecule, including modifications like methylation. Direct sequencing provides valuable insights into post-transcriptional modifications, making it particularly useful for gene regulation studies.
4. Real-Time Analysis
- PacBio: Unlike Nanopore, PacBio doesn’t offer real-time analysis. Sequencing occurs in multiple passes, which can extend data generation times. However, PacBio compensates for this by producing highly accurate, high-quality reads, making it ideal for research requiring precision over speed.
- Nanopore: Nanopore’s real-time sequencing is one of its standout features. It enables immediate analysis as data is generated, allowing for quick decision-making. This is beneficial in scenarios such as clinical diagnostics or field-based research, where fast results are crucial. Real-time data analysis is also great for continuous monitoring and quality control during sequencing.
5. Throughput
- PacBio: PacBio provides high-throughput sequencing. However, due to its longer read lengths and the HiFi sequencing method, the process can be slower compared to Nanopore. The multiple sequencing passes required for greater accuracy can lead to longer processing times, though the data quality remains top-notch.
- Nanopore: Nanopore boasts high-throughput sequencing with faster data generation. The simplicity of its workflow (no cDNA conversion or amplification) allows for rapid processing. However, this speed may come at the expense of accuracy, especially with longer reads, requiring additional error correction steps for high-quality results.
6. Application Suitability
- PacBio: PacBio is best suited for applications that demand high accuracy, such as transcriptome analysis, de novo genome assembly, structural variant detection, and haplotype phasing. Its HiFi sequencing technology is particularly beneficial for detecting rare variants and resolving complex genomic regions.
- Nanopore: Nanopore excels in applications where real-time sequencing and portability are crucial. It’s commonly used in field-based research, clinical diagnostics, and studies requiring rapid data analysis. While its accuracy might not match PacBio’s, Nanopore’s ability to directly sequence RNA and handle high-throughput applications makes it perfect for epigenetic research and transcriptome analysis.
Both PacBio and Nanopore bring distinct advantages to RNA sequencing. PacBio is the go-to for high-accuracy applications, such as structural variant detection and gene expression analysis. Its long, accurate reads make it ideal for complex transcriptomic studies.
In contrast, Nanopore offers unique real-time sequencing capabilities and portability, making it suitable for rapid analysis and field-based research. The choice between these two technologies will depend on your project’s focus—whether you prioritize speed and portability or require high precision and depth.
Applications in Research
RNA sequencing technologies like PacBio and Nanopore are changing genomics research. With long-read sequencing from PacBio and real-time capabilities from Nanopore, these tools are pushing the boundaries of large-scale projects, from cancer research to tracking infectious diseases.
Application of PacBio Technology
For example, PacBio was heavily involved in the “Telomere-to-Telomere” (T2T) assembly of the human genome, which aimed to produce a complete, error-free reference of the human genome. The gapless human genome sequence includes 3.055 billion base pairs of DNA, offering an unprecedented level of detail. It fills in previously missing sections, particularly in highly repetitive regions that traditional sequencing methods had been unable to resolve. This involved HiFi sequencing to assemble long, accurate reads that could capture previously overlooked regions, such as repetitive sequences and structural variations.
PacBio’s technology allowed for a high-quality, comprehensive human genome map, an essential resource for genomics research. HiFi sequencing was used to fill gaps that traditional short-read sequencing could not address, helping complete the human genome with previously unavailable telomeric and centromeric regions.
PacBio’s long-read sequencing has also been applied in projects like The Cancer Genome Atlas (TCGA) to study the genetic variations in cancer. The Cancer Genome Atlas, a collaborative initiative between the National Cancer Institute (NCI) and the National Human Genome Research Institute, molecularly characterized over 20,000 cancers and matched normal samples across 33 cancer types. Launched in 2006, TCGA brought together researchers from various disciplines to generate over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data.
By identifying structural variants and mutations in large tumor datasets, PacBio’s sequencing technologies provide valuable insights into cancer genomics. Its long reads and HiFi sequencing have been essential in uncovering gene rearrangements and transcript isoforms in various cancer types.
Using PacBio, researchers have been able to track genetic changes in tumors, helping to improve our understanding of cancer’s genetic foundations and contributing to the development of more tailored, effective cancer treatments.
Application of Nanopore Technology
During the COVID-19 pandemic, Oxford Nanopore launched the Midnight Kit, designed for rapid and cost-effective sequencing of SARS-CoV-2 across various throughput scales. The MinION, compatible with Oxford Nanopore sequencing devices, enables genomic surveillance with minimal hands-on time, making it easy to automate and scale for large-scale projects. It allows for the sequencing of tens to thousands of samples with costs as low as $9.55 per sample.
The Midnight Kit supports workflows that can process 12 samples in 7 hours or scale up to 480 genomes in under a day. This flexibility, paired with the ARTIC method, made it a valuable tool for COVID-19 surveillance in diverse laboratory settings. Nanopore’s real-time sequencing capabilities were essential for providing immediate genomic data to inform public health strategies and vaccine development.
Now, it is clear that PacBio shines in large-scale projects that require high accuracy and the ability to resolve complex genomic regions, making it ideal for projects like human genome assembly and cancer genomics.
Nanopore, on the contrary, excels in real-time applications, especially when portability and speed are essential, as seen in the rapid sequencing of SARS-CoV-2 and AMR surveillance. Its direct RNA sequencing and quick turnaround times make it valuable for field-based research and real-time data analysis.
To Sum Up!
Both PacBio and Nanopore bring unique advantages to RNA sequencing. PacBio offers high-accuracy, long-read sequencing ideal for complex genomic studies, while Nanopore excels in real-time, portable sequencing for fast, flexible analysis.
PacBio’s HiFi sequencing is best for precise, high-quality data, while Nanopore’s direct RNA sequencing and scalability make it perfect for rapid, on-site research. The choice between these technologies ultimately depends on your research goals—whether you prioritize accuracy and depth or speed and portability.
For those seeking a cost-effective, high-quality RNA sequencing solution without the complexity, Biostate AI offers a powerful alternative. With its affordable Total RNA sequencing, Biostate AI streamlines the sequencing process from sample collection to data analysis, allowing researchers to focus on their scientific discoveries. Whether you’re studying mRNA, IncRNA, miRNA, or other RNA types, Biostate AI delivers comprehensive insights with minimal effort and cost.
Get a quote now for your experiments and take advantage of Biostate AI’s affordable, high-quality RNA sequencing solutions that empower your research without breaking your budget.
