Did you know? According to the International Agency for Research on Cancer (IARC), every minute, four women are diagnosed with breast cancer, and one dies from the disease.
Despite significant advancements in cancer research, breast cancer remains highly heterogeneous, presenting challenges in treatment and resistance to therapies. Traditional methods, like bulk sequencing, fall short in capturing this complexity, limiting the precision of treatments.
scRNA-seq holds the key to unlocking new therapeutic targets and overcoming drug resistance by providing insights at the cellular level, ultimately improving outcomes for millions of women.
In this blog, we will explore how scRNA-seq in breast cancer is shaping the future of personalized medicine and its potential to reform breast cancer treatment strategies.
TL;DR
- Unlike bulk sequencing, scRNA-seq captures intra-tumor heterogeneity, offering a clearer view of tumor progression and enabling more personalized medicine.
- scRNA-seq helps identify therapeutic targets and drug resistance mechanisms, all crucial for improving treatment strategies and patient outcomes.
- scRNA-seq provides precise data on tumor microenvironments, cancer stem cells, metastasis, and immune interactions, contributing to a better understanding and potential therapies.
- An affordable, fast, and high-quality RNA-Seq with AI-driven analysis is an ideal choice for researchers looking to advance their scRNA-seq in breast cancer studies.
What is sc-RNA-seq?
Single-cell RNA sequencing (scRNA-seq) is a powerful technology that allows researchers to examine gene expression profiles at the level of individual cells.
scRNA-seq vs. Bulk RNA Sequencing
Unlike traditional bulk RNA sequencing, scRNA-seq captures the unique transcriptomic information of each cell.
- Resolution: scRNA-seq provides individual cell data, revealing rare subpopulations; bulk RNA-seq averages signals from all cells, masking variability.
- Heterogeneity: scRNA-seq captures intra-tumor heterogeneity and cell-state transitions, while bulk RNA-seq fails to highlight these differences.
- Gene Expression Variability: scRNA-seq detects gene expression variability, crucial for understanding cancer dynamics, whereas bulk RNA-seq averages out key details.
By analyzing the RNA of each individual cell, scRNA-seq reveals hidden insights into cellular heterogeneity, regulatory mechanisms, and disease dynamics.
scRNA-seq Protocols and Methodologies
Single-cell RNA sequencing (scRNA-seq) protocols have rapidly evolved, offering researchers a range of methods tailored for sensitivity, throughput, and cost-effectiveness.
The core workflow typically involves isolating individual cells from breast tumor tissue, extracting RNA, reverse-transcribing it into cDNA, amplifying the genetic material, and sequencing it to generate a transcriptomic profile for each cell.

Key steps and considerations include:
- Single-cell isolation: Techniques such as microfluidics, fluorescence-activated cell sorting (FACS), and droplet-based systems (e.g., 10x Genomics Chromium) are commonly used to separate viable single cells from complex tumor samples. Recent advances have made these methods more automated and scalable, improving the accuracy and efficiency of cell capture.
- Library preparation: Protocols like Smart-seq, Smart-seq2, Quartz-Seq, and CEL-seq enable precise capture and amplification of mRNA from individual cells, with each cell receiving a unique barcode for downstream data analysis.
- Sequencing: High-throughput sequencing platforms, typically Illumina-based, are used to read the barcoded cDNA libraries, generating large datasets that capture the transcriptome of thousands of cells in parallel.
- Data analysis: Bioinformatics pipelines process raw sequencing data, perform quality control, normalize expression levels, and cluster cells based on gene expression patterns. This enables the identification of rare cell types, mapping of tumor heterogeneity, and exploration of cell-cell interactions within the breast cancer microenvironment.
Recent innovations in scRNA-seq methodologies include integration with multiomics approaches, spatial transcriptomics, and liquid biopsy applications, further enhancing the ability to dissect tumor complexity and monitor disease progression non-invasively.
Applications of scRNA-seq in Breast Cancer Research
scRNA-seq provides new perspectives for understanding the mechanisms behind tumor therapy, drug resistance, and metastasis in breast cancer, highlighting its impact on the comprehension of breast cancer biology and its potential for guiding personalized treatment strategies.
- Tumor Heterogeneity Analysis and Cellular Composition
Breast tumors are complex ecosystems comprising malignant cells, stromal components, and diverse immune populations, each with distinct phenotypic states. Single-cell RNA sequencing (scRNA-seq) has emerged as a transformative tool for dissecting this heterogeneity at unprecedented resolution.
Identification of Molecular Subtypes and Cellular Diversity
scRNA-seq on breast tumors revealed that the cellular composition varies significantly between patients and molecular subtypes, with important implications for treatment selection.
- A breakthrough study utilized scRNA-seq to identify CD44+/ALDH2+/ALDH6A1+ breast cancer stem cells (BCSCs) in primary tumors.
- Key Findings:
- BCSCs represent a distinct cellular population with unique metabolic characteristics
- These cells serve as precursors for metastatic disease
- NECTIN2-TIGIT-mediated interactions promote immune escape and lymph node metastasis
- Key Findings:
- In Triple-Negative Breast Cancer (TNBC), an especially aggressive subtype, single-cell profiling has identified five epithelial clusters within primary tumors.
- Notably, cluster 2, marked by a high-cycling, luminal progenitor-like (LP) signature, correlates with worse survival, highlighting both potential cells of origin and prognostic biomarkers.
This approach extends to other subtypes (e.g., luminal A/B, HER2+), where scRNA-seq surpasses bulk profiling by resolving molecular heterogeneity critical for precision therapy.
Identification of Intra-tumor Heterogeneity
scRNA-seq studies have revealed that even within individual tumors, cancer cells exhibit substantial heterogeneity.
- A comprehensive analysis of 96,796 single cells from 15 paired samples of primary tumors and lymph node metastases was conducted.
- This study identified multiple cancer cell subclusters, including breast cancer stem cells (BCSCs) that exist only in primary tumors and evolve into metastatic clusters.
- This finding has profound implications for understanding metastatic progression and developing anti-metastatic therapies.
Studies using 10x Visium spatial transcriptomics have revealed that tumor heterogeneity is not randomly distributed but exhibits specific spatial patterns that influence cellular interactions and treatment responses.
- Mapping the Tumor Microenvironment (TME) at Single-Cell Resolution
The breast TME is a dynamic interplay of malignant and non-malignant cells. scRNA-seq has comprehensively mapped the following cell types and functions:
| Cell Type Category | Specific Cell Type | Key Characteristics/Role in Breast Cancer (as revealed by scRNA-seq) |
| Epithelial | Luminal Hormone-Responsive Epithelial Cells | Abnormal morphology, proliferation, and function in cancer; sensitive to estrogen/progesterone; involved in proliferation/differentiation. |
| Epithelial | Luminal Secretory Epithelial Cells | Involved in milk synthesis/secretion; abnormal proliferation/differentiation in disease. |
| Epithelial | Basal Myoepithelial Cells | Provide structural support, regulate tissue tension, participate in immune defense, and secrete growth factors. |
| Stromal | Endothelial Cells | Line blood/lymphatic vessels; maintain integrity/permeability; regulate flow; respond to hormones. |
| Stromal | Mesenchymal Cells (MSCs) | Provide structural support; regulate microenvironment; accelerate cancer cell proliferation/invasion in TME. |
| Stromal | Perivascular Cells | Form blood vessel walls; regulate contraction/dilation; participate in vessel repair. |
| Stromal | Adipocytes | Primarily store fat, and provide energy reserves. |
| Stromal | Cancer-Associated Fibroblasts (CAFs) | Diverse subpopulations (e.g., inflammatory-like, matrix); regulate the extracellular matrix, associated with T-cell dysfunction. |
| Immune | Macrophages | Phagocytosis, immune regulation; show polarization shifts (pro-inflammatory to pro-tumorigenic) in metastasis. |
| Immune | T Cells | Maintain immune homeostasis; exhibit continuous activation states rather than discrete subtypes; potential immunotherapy targets (e.g., CD8+ TRM). |
| Immune | B Cells | Produce antibodies; provide immune memory; involved in tumor development and anti-tumor immunity. |
| Immune | Natural Killer (NK) Cells | Directly attack infected or tumor cells; can be reprogrammed by cancer cells to promote tumors. |
| Immune | Mast Cells | Trigger inflammatory/immune responses; involved in breast remodeling. |
- CTCs and Stromal Diversity
scRNA-seq of circulating tumor cells (CTCs) provides a window into metastatic potential. A pioneering study used scRNA-seq to analyze circulating tumor cells (CTCs) from breast cancer patients.
The research identified two distinct CTC populations: CTC-1 cells with high proliferative capacity and estrogen responsiveness, and CTC-2 cells with epithelial-mesenchymal transition (EMT) characteristics and enhanced metastatic potential.
Clinical Implications:
- CTC-1 cells may be targetable with anti-endocrine therapy
- CTC-2 cells represent a more aggressive, therapy-resistant population
- Differential CTC characterization could guide treatment decisions
Stromal cell profiling has uncovered diverse subpopulations of cancer-associated fibroblasts (CAFs) and perivascular-like (PVL) cells with distinct localizations, surface markers, and matrix-modulating functions.
Notably, inflammatory CAF and differentiated PVL gene signatures are associated with T cell dysfunction and exclusion, pointing to a critical stromal-immune axis in immune escape and therapy resistance.
- Mapping Drug Resistance Mechanisms and Targetable Pathways
Relapse and metastasis in breast cancer are often driven by drug-resistant cell populations, either pre-existing or acquired. scRNA-seq enables high-resolution mapping of phenotypically distinct cancer cells, revealing subtle differences in drug sensitivity that are often missed by bulk sequencing.
Cellular Plasticity and Transcriptional Reprogramming in Resistance
Drug resistance is not limited to clonal selection. A major mechanism involves cellular plasticity, which is the ability of tumor cells to adopt alternative phenotypes that support survival.
Integrated analysis using scRNA-seq, snRNA-seq, and scDNA-seq shows that resistance is frequently driven by post-treatment transcriptional reprogramming, rather than fixed genomic alterations.
While resistant clones may pre-exist, many cells acquire resistant phenotypes through dynamic gene expression changes following drug exposure.
Key upregulated pathways in chemoresistant cells include:
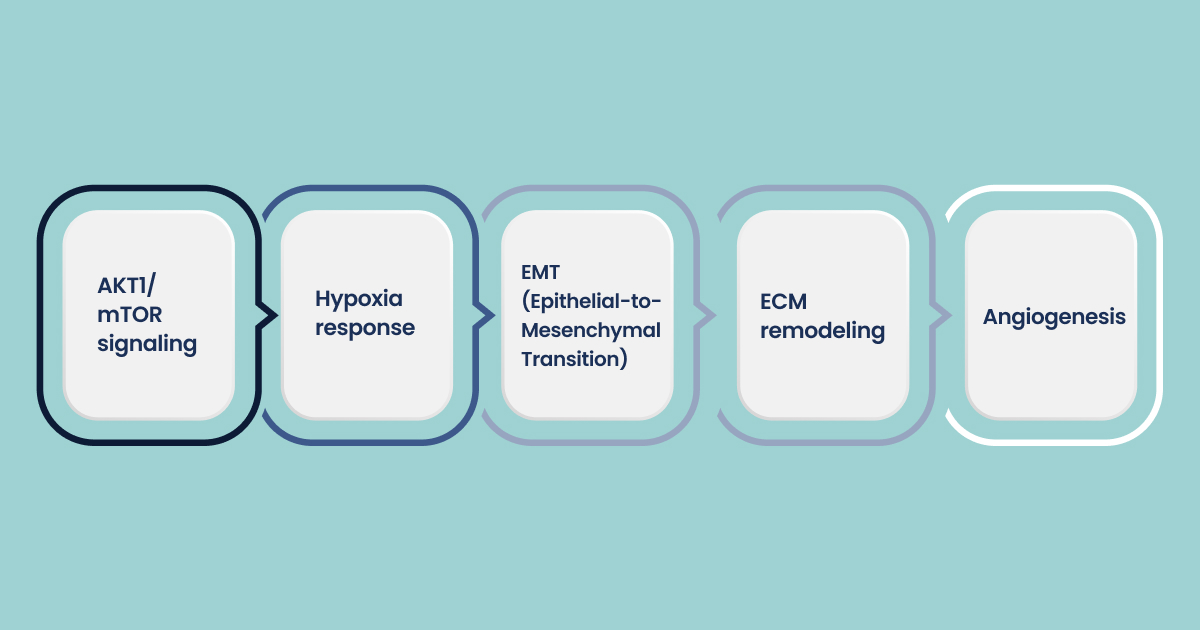
- AKT1/mTOR signaling
- Hypoxia response
- EMT (Epithelial-to-Mesenchymal Transition)
- ECM remodeling
- Angiogenesis
These pathways are not incidental. They are directly involved in therapeutic failure and can serve as intervention points.
Recent tools like SSDA4Drug infer drug-resistant phenotypes directly from scRNA-seq data, even with minimal labeled inputs. These models track temporal changes in drug sensitivity and map resistance across cancer cell lineages.
Target Discovery and Therapeutic Implications
scRNA-seq enables identification of cell states and transcriptomic programs linked to resistance, creating an opportunity for targeted therapeutic design. Gene expression profiles associated with EMT, stemness, proliferation, and recurrence offer precise targets for next-generation drug development.
The dual model of resistance, pre-existing versus acquired, has clear implications for therapy:
- Pre-existing resistance: Early detection and molecular stratification are key. scRNA-seq can identify resistant clones before treatment.
- Acquired resistance: Requires tracking of transcriptional shifts post-therapy. Targeting reprogramming pathways can delay or reverse resistance.
Critically, these processes are not necessarily driven by new mutations. Resistance is often epigenetically regulated and mediated through transcription factor networks, chromatin remodeling, and non-coding RNA activity. Therefore, therapies targeting only oncogenic mutations are insufficient.
Combining standard treatments with inhibitors of reprogramming pathways (e.g., mTOR inhibitors or EMT blockers) or differentiation-inducing agents could re-sensitize resistant cells. Moreover, single-cell profiling of patient samples before and after treatment offers a direct method to measure response evolution and adapt therapy in real-time.
- Advancing the Understanding of Metastasis and Disease Progression
Metastasis, the spread of cancer cells from the primary tumor to distant sites, is the leading cause of breast cancer-related deaths. Traditionally, studying the intricate processes involved in metastasis has been challenging due to the complexity and heterogeneity of tumors. scRNA-seq has redefined this field by providing a high-resolution, single-cell view of the tumor ecosystem.
The Role of Epithelial-to-Mesenchymal Transition (EMT) in Disease Progression
One of the key processes associated with metastasis is Epithelial-to-Mesenchymal Transition (EMT), which facilitates cancer cell invasion, dissemination, and resistance to therapies.
- However, EMT is not a fixed state but a continuous progression through different cell stages, making it challenging to study using traditional bulk RNA sequencing methods.
- scRNA-seq allows for a more nuanced understanding by capturing the various intermediate stages of EMT at single-cell resolution.
- The scPrognosis method has been specifically designed to leverage scRNA-seq data from EMT processes to identify prognostic signatures in breast cancer.
- scPrognosis analyzes gene expression variations during EMT, integrating key measures such as the absolute deviation in gene expression, the differentiation across different stages of EMT, and the role of genes within the dynamic co-expression network during this transition.
By reconstructing the pseudotemporal trajectory of cells undergoing EMT, scPrognosis enables researchers to track the progression of EMT, providing critical insights into its role in cancer progression and therapy resistance.
Comparative Analysis of Primary and Metastatic Tumor Landscapes
A major advantage of scRNA-seq is its ability to compare primary and metastatic tumor landscapes in great detail.
- Studies using scRNA-seq to analyze estrogen receptor-positive (ER+) breast cancer samples from both primary and metastatic sites have revealed significant differences in the tumor microenvironment (TME).
- While the main cell populations, such as malignant cells, T cells, and myeloid cells, were found in both tumor types, minor cell subtypes exhibited notable variations between primary and metastatic tumors.
Additionally, scRNA-seq revealed differences in gene regulatory signatures between primary and metastatic cells.
- Primary tumors showed higher activity in transcription factors related to tumor development, apoptosis, and immune differentiation, such as ETS2, EPAS1, and BATF.
- In contrast, metastatic cells exhibited heightened activity in factors like HOXC13, GATA2, and IRF9, which are linked to poor prognosis and metastatic spread.
These findings provide potential targets for therapeutic intervention aimed at inhibiting the transcriptional programs driving metastasis.
- Biomarker Discovery and Personalized Medicine
By examining individual cells within both primary and metastatic tumors, scRNA-seq enables the identification of distinct gene expression profiles that are specific to various cell types, including both malignant and non-malignant cells.
One of the most significant contributions of scRNA-seq in breast cancer research is its ability to uncover gene regulatory signatures that differentiate primary tumors from metastatic cells. By analyzing these signatures, researchers can identify key transcription factors that are active in either the primary or metastatic stages of cancer.
- For instance, transcription factors play essential roles in regulating processes like tumor growth and immune responses, making them critical for understanding how primary tumors evolve and interact with the immune system.
- The identification of these transcriptional signatures provides new potential biomarkers for predicting the risk of metastasis and therapeutic outcomes.
By analyzing scRNA-seq data from a patient’s primary tumor, clinicians can also determine which transcriptional programs are driving disease progression and tailor treatments accordingly.
- For instance, if a patient’s tumor exhibits high activity in ETS2 or BATF, therapies that target these pathways might be more effective in halting tumor growth.
- On the other hand, if HOXC13 or GATA2 is more active in the metastatic cells, personalized therapies designed to inhibit these factors could help in preventing further metastasis.
This precision enables more effective, individualized treatments and helps overcome challenges such as drug resistance and metastasis, offering hope for improved outcomes in cancer therapy.
Technical Considerations and Challenges of Breast Cancer Research with scRNA-seq

Here are the challenges associated with the widespread adoption and clinical translation of single-cell sequencing (SCS) technologies:
- High Cost and Technical Difficulties: Despite decreasing costs and streamlined protocols, SCS remains expensive and technically challenging, limiting its widespread use in clinical settings.
- Data Integration: Integrating large, diverse datasets from single-cell experiments is a significant hurdle, requiring advanced bioinformatics tools capable of converting these data into actionable insights for treatment strategies.
- Working with Degraded Samples: Degraded biological samples, particularly those from clinical settings, can lead to low-quality RNA, making it difficult to obtain reliable scRNA-seq data.
- Adapting Bioinformatics Tools: Existing bioinformatics tools, primarily developed for developmental biology, need adaptation to disease contexts, such as breast cancer, presenting a challenge in translating findings into clinical applications.
- Doublet Contamination: Doublet contamination, where two cells are mistakenly sequenced as one, can impact the quality and accuracy of scRNA-seq data, complicating the interpretation of cellular populations.
These issues not only require advanced technical expertise but also consume valuable time and resources, making it difficult for researchers to focus solely on their studies. Here’s where Biostate AI shines.
How Biostate AI Can Offer You Efficient RNA-Sequencing Analysis
Biostate AI provides an efficient solution to these obstacles, offering a comprehensive, end-to-end RNA sequencing service. Our platform combines AI-driven analytics with high-quality sequencing at an affordable price, making it a perfect fit for researchers seeking both cost-efficiency and precision in their RNA sequencing projects.
Our offerings:
- Unbeatable Pricing: High-quality sequencing results starting at just $80/sample.
- Rapid Turnaround: Receive your results in 1–3 weeks.
- Complete Transcriptome Insights: Comprehensive RNA-Seq covering both mRNA and non-coding RNA.
- AI-Driven Analysis: Access powerful, intuitive insights with OmicsWeb AI.
- Minimal Sample Requirement: Process samples as small as 10µL blood, 10ng RNA, or 1 FFPE slide.
- Low RIN Compatibility: Compatible with RNA samples having a RIN as low as 2 (vs the typical ≥5).
- OmicsWeb: A robust platform combining data storage and automated analysis workflows, ready for AI-driven insights.
- Disease Prognosis AI: AI-powered disease prediction and therapy guidance through Biobase, using RNA data to predict drug toxicity and therapy efficacy.
With Biostate AI, we revolutionize RNA sequencing by offering both affordability and comprehensive, AI-enhanced analytics, ensuring our research achieves the highest standards of quality and insight, regardless of sample type or condition.
Final Words!
scRNA-seq in breast cancer is offering researchers a detailed view of the tumor’s cellular composition. This allows a deeper understanding of cancer heterogeneity, drug resistance, and metastasis.
By examining gene expression at the level of individual cells, scRNA-seq in breast cancer enables the identification of distinct cellular populations that drive the disease, paving the way for more precise, targeted treatments.
To support the growing need for high-quality scRNA-seq data, Biostate AI offers comprehensive RNA-Seq, covering both mRNA and non-coding RNA, with AI-driven analysis to facilitate data interpretation.
Whether you are working with small sample sizes or challenging RNA conditions, our service is designed to ensure reliable, accurate results in as little as 1–3 weeks, starting at just $80/sample.
Get your quote today and see how we can help you advance your scRNA-seq research with precise, cost-effective solutions that make your work easier and faster.
FAQs
- How can single-cell RNA sequencing uncover the role of non-cancerous cells in breast tumor progression?
Beyond cancer cells, scRNA-seq reveals the diverse populations of stromal, immune, and fibroblast cells within the tumor microenvironment, elucidating their dynamic interactions that influence tumor growth, metastasis, and immune evasion. Understanding these non-malignant players is crucial for developing combination therapies targeting both the tumor and the microenvironment.
- Can scRNA-seq help predict breast cancer patient prognosis beyond traditional clinical markers?
Yes, scRNA-seq identifies novel cell-type-specific gene expression signatures and rare subpopulations linked to aggressive disease and poor outcomes, offering prognostic biomarkers that outperform conventional histopathological and molecular subtyping methods.
- What ethical and data privacy challenges arise from single-cell sequencing data in breast cancer research?
The granular nature of scRNA-seq data raises concerns about patient privacy, data sharing, and consent, especially as datasets grow and integrate with clinical records. Addressing these issues requires robust governance frameworks to protect patient identity while enabling scientific collaboration.
- How might integrating single-cell RNA sequencing with spatial transcriptomics transform breast cancer research?
Combining scRNA-seq with spatial transcriptomics preserves tissue architecture, allowing researchers to map gene expression to precise locations within tumors. This integration reveals spatial heterogeneity and cell-cell communication networks, offering deeper insights into tumor biology and therapeutic vulnerabilities.
